Fundamental Features of Prion Proteins
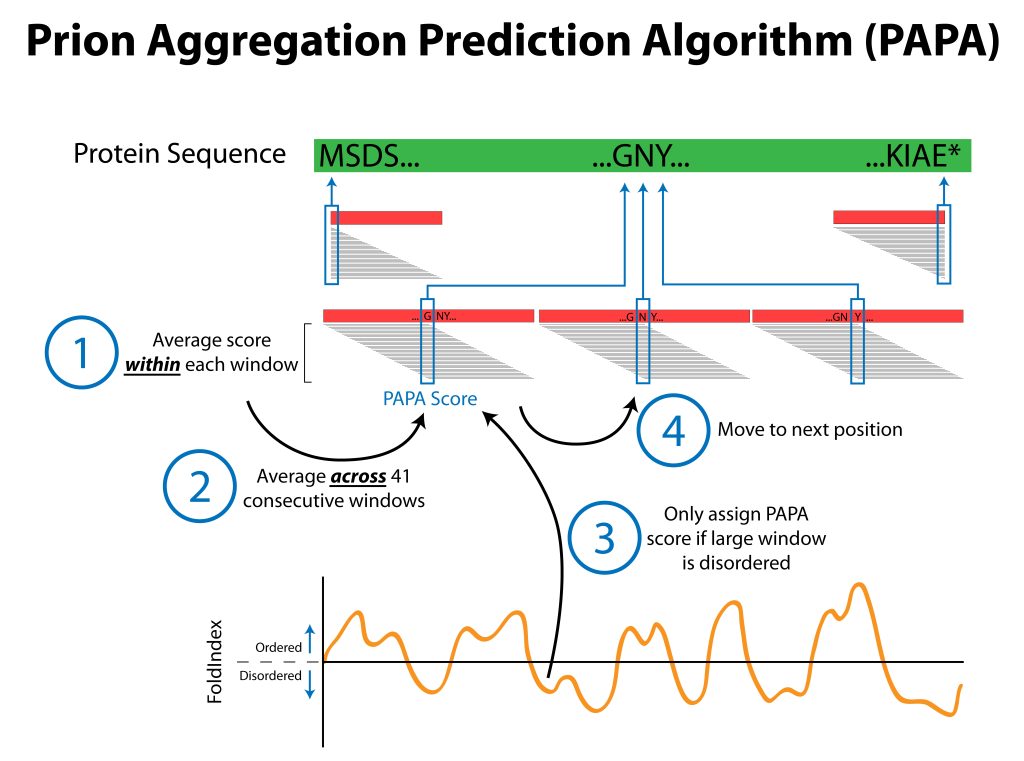
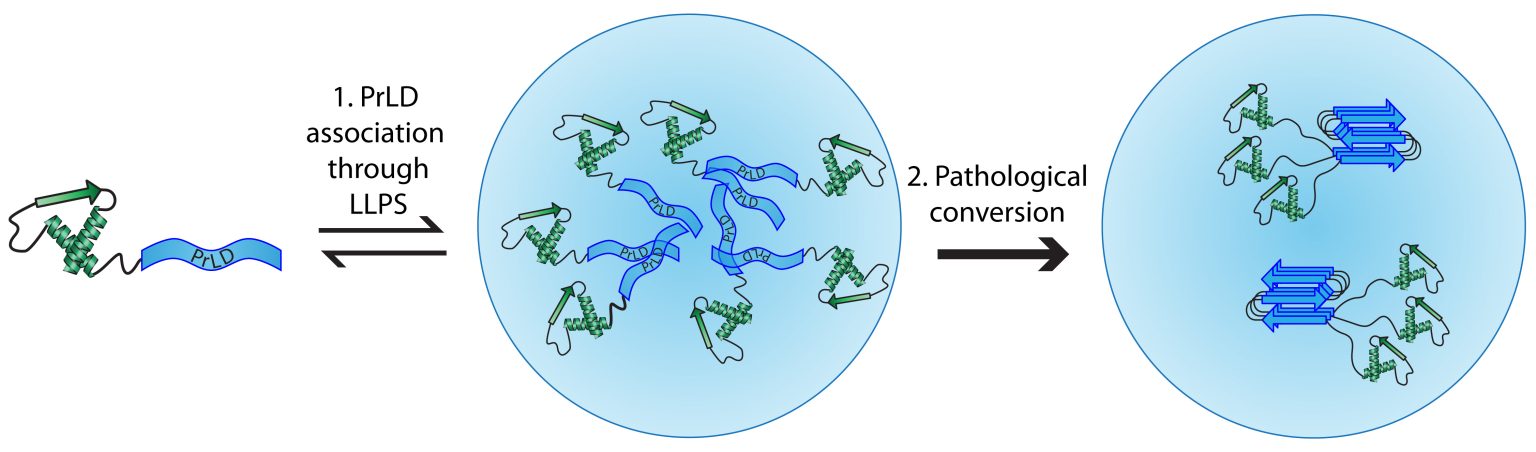
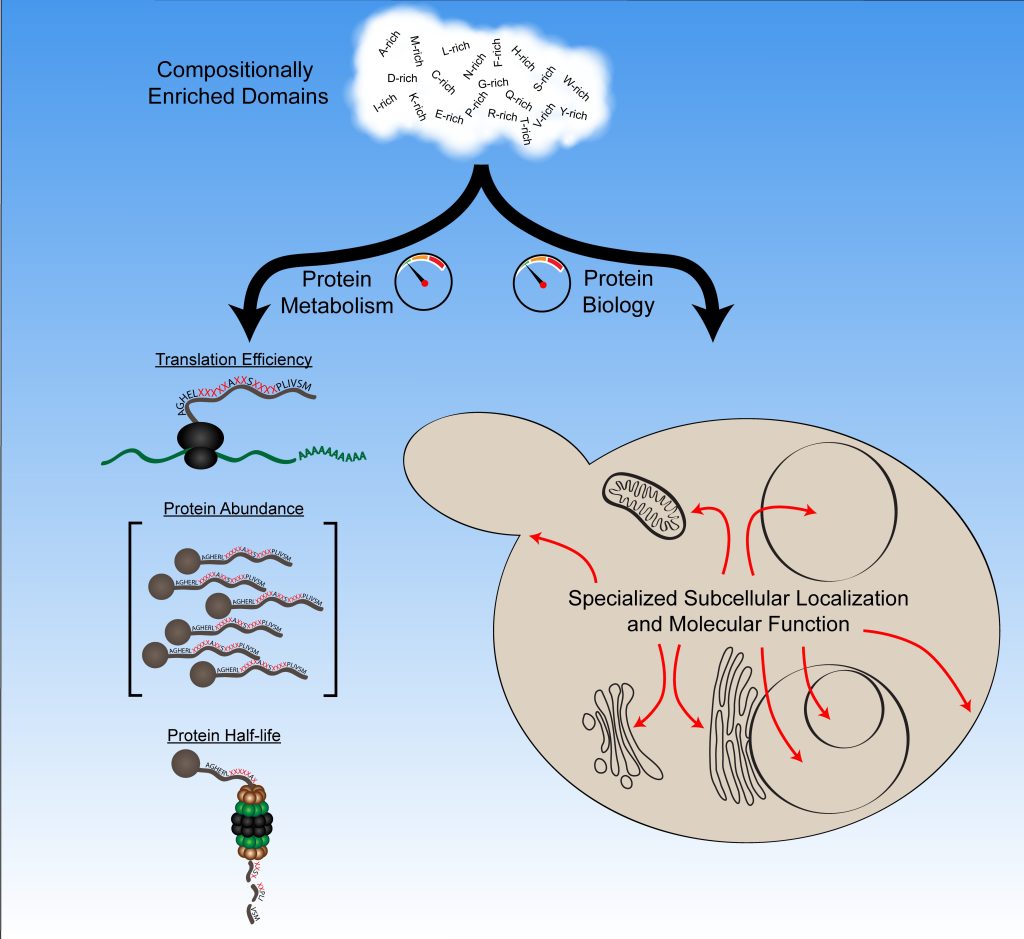
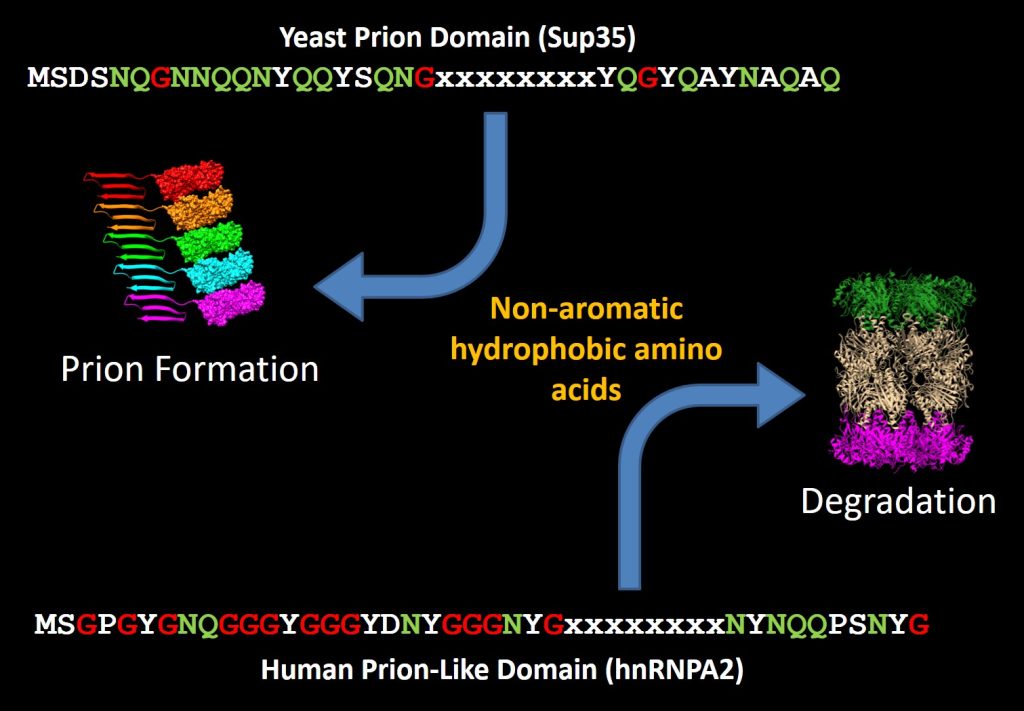
Our lab uses budding yeast to study the basic steps involved in the formation of infectious protein aggregates (“prions”). Specifically, we are interested in understanding the fundamental features of prion proteins. In yeast, a growing list of proteins have the demonstrated ability to form prions. Known prion proteins often contain strikingly Q/N-rich prion domains. However, while this is a common feature among prion proteins, it is not sufficient to predict prion activity. Therefore, using a large-scale genetic screen, we have developed a bioinformatic method to predict prion propensity.